


Next: Search for genes of
Up: Start by constructing a
Previous: Start by constructing a
Contents
Using predefined gene list(s)
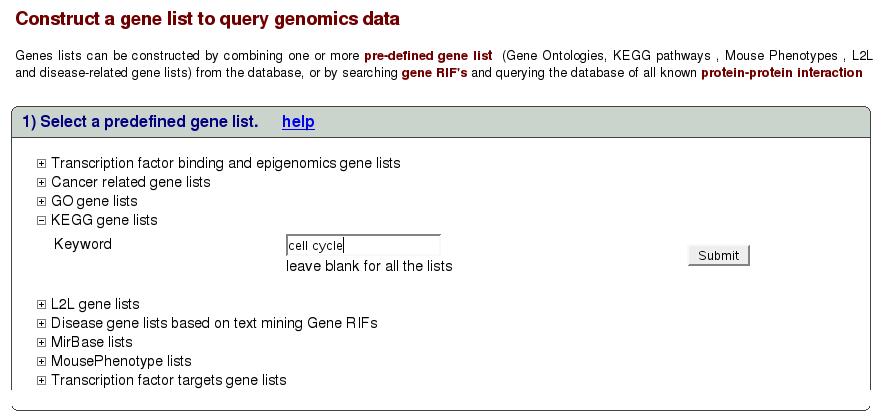
Figure![[*]](crossref.png) shows the interface to select
a predefined gene list. Clicking on ``Gene List'' tab in the left
menu would get this page. The lists are organized in different categories
and we are constantly adding new lists and categories. Let's say we
are interested in gene lists in category ``KEGG'' with keywords
``cell cycle''. Click on the link ``KEGG gene list'' to expand
the search box as shown below. Type cell cycle in the text box and
click submit.
shows the interface to select
a predefined gene list. Clicking on ``Gene List'' tab in the left
menu would get this page. The lists are organized in different categories
and we are constantly adding new lists and categories. Let's say we
are interested in gene lists in category ``KEGG'' with keywords
``cell cycle''. Click on the link ``KEGG gene list'' to expand
the search box as shown below. Type cell cycle in the text box and
click submit.
Figure:
Search a predefined gene list
|
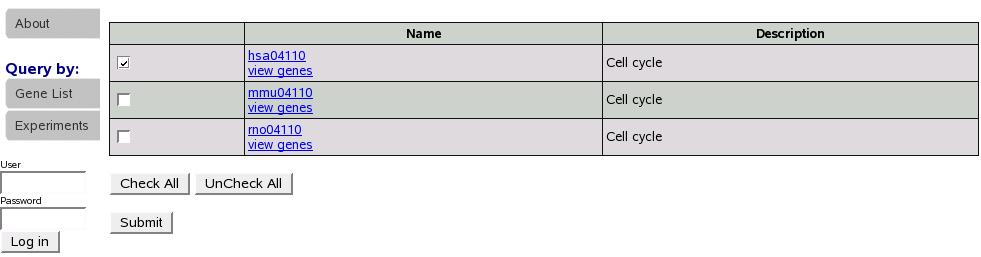
This takes us to the following screen (figure![[*]](crossref.png) ).
Here we see a list of gene list returned for the keywords. Select
one or multiple lists using the check boxes and click submit.
).
Here we see a list of gene list returned for the keywords. Select
one or multiple lists using the check boxes and click submit.
Figure:
Select gene list
|
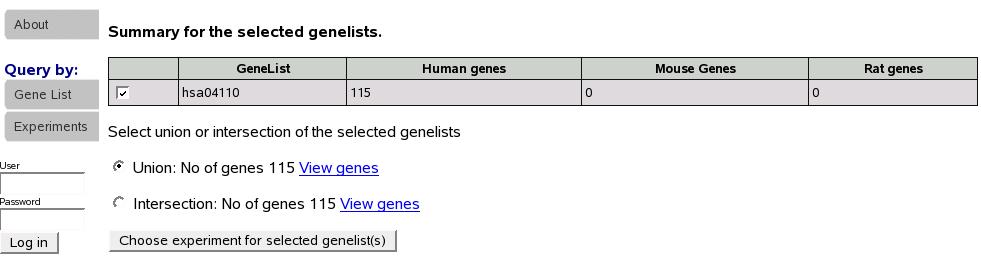
Now we see summary for the submitted lists. Note that below the summary
table, we are asked to select union or intersection of the gene lists
selected. In this case, because we submitted only one gene list, both
union and intersection are identical. Let's select union and proceed
to select an experiment for analysis.
Figure:
Summary of gene list
|
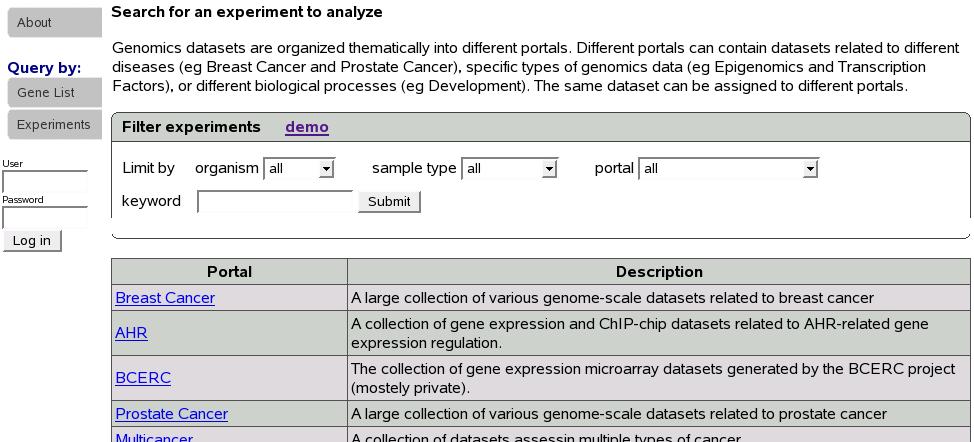
Experiments are organized into different portals. If we know the portal
the experiment of our interest belongs to, we can click on the portal
name to list all the experiments in that portal. Since this is not
the case most times, a search function is provided to look for experiments
of our interest. In the ``Filter experiments'' box shown below,
select organism, sample type, portal of your interest and type in
keyword. Keyword could be left blank.
Figure:
Select or search for experiments
|
Let's click on portal name ``Breastcancer'' to proceed with our
example.
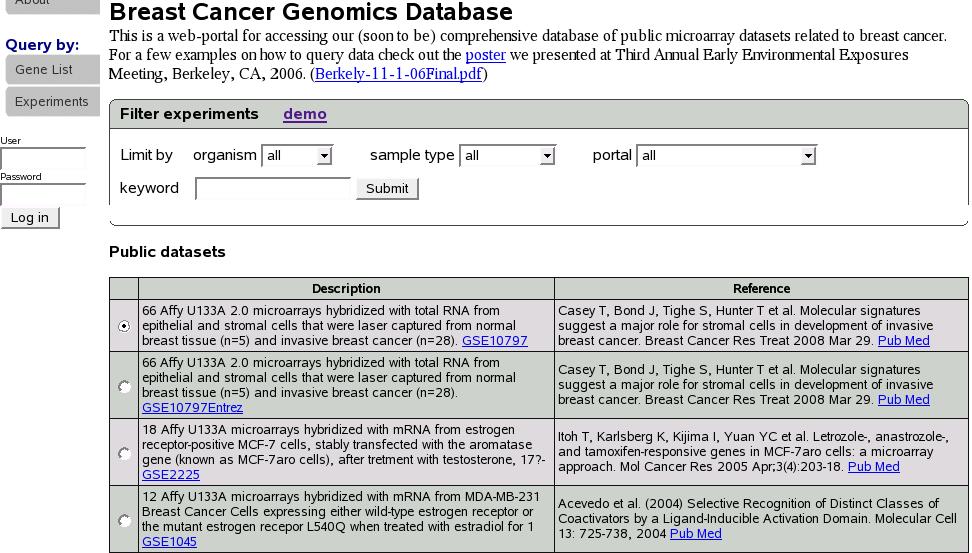
Figure![[*]](crossref.png) is a part of the screen showing
experiments in the portal Breastcancer. If we had used the search
function to look for an experiment instead, a similar screen would
be shown. Below you would see a list of experiments. Let's select
first experiment (GSE10797) and scroll down and click submit.
is a part of the screen showing
experiments in the portal Breastcancer. If we had used the search
function to look for an experiment instead, a similar screen would
be shown. Below you would see a list of experiments. Let's select
first experiment (GSE10797) and scroll down and click submit.
Figure:
show experiments
|
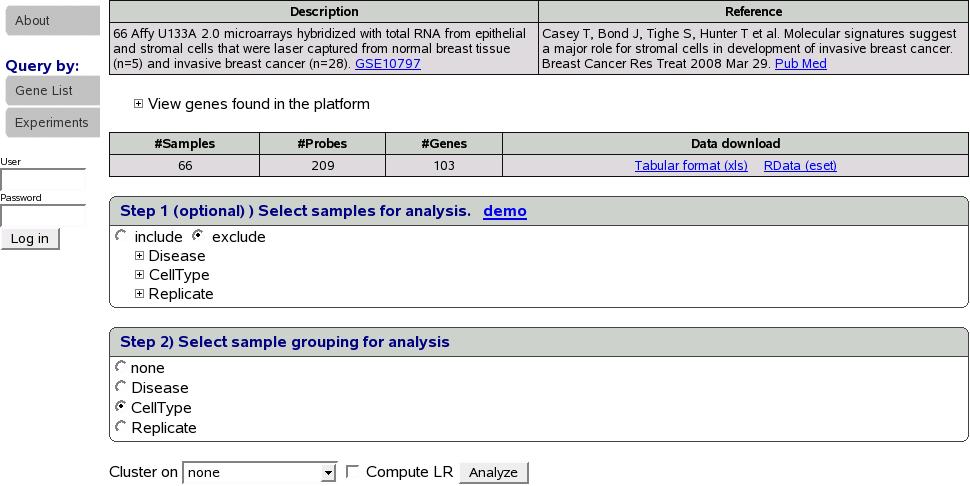
At this point, data is retrieved from database for the selected experiment
and gene list(s) as shown in figure![[*]](crossref.png) .
.
Figure:
Get data
|
One could download the data for his/her own analysis either as a tab
delimited file or an eset for analysis in R. In this example, we have
66 samples, 209 probes and 103 genes. If we want to analyze only a
subset of samples, we could select samples using the ''Step 1''
shown above. This step is optional and default is to select all the
samples. Next we select a sample grouping for the analysis. We could
choose to cluster on genes, samples, both genes and samples or none
using the combo box shown above. Check the box ``Compute LR''
to compute predictive ability pvalue. Let's select ``CellType''
sample grouping, leave step 1 as it is to select all the samples,
cluster on ``none'' and click Analyze button.
Figure:
Results
|
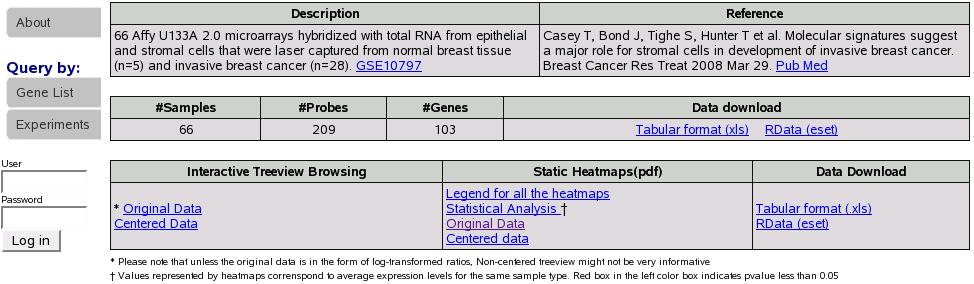
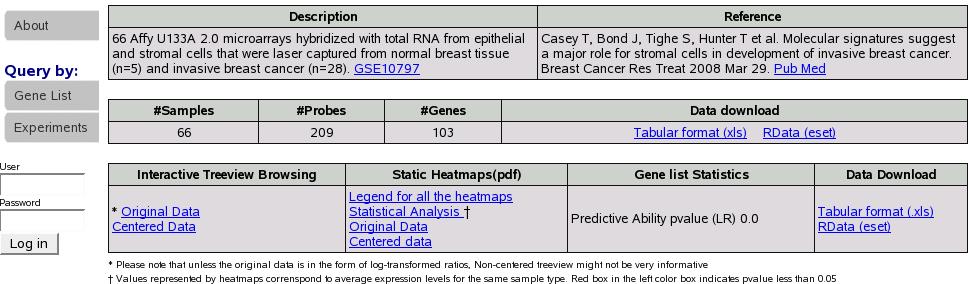
If we had checked the compute LR box we would see an additional column
``Gene list Statistics'' with the computed pvalue in the results
tables as shown in figure![[*]](crossref.png) .
.
Figure:
Results with LR
|
Figure ![[*]](crossref.png) depicts a typical summary table details
of which can be found in Section Interpreting Results'.
depicts a typical summary table details
of which can be found in Section Interpreting Results'.



Next: Search for genes of
Up: Start by constructing a
Previous: Start by constructing a
Contents
![[*]](crossref.png) shows the interface to select
a predefined gene list. Clicking on ``Gene List'' tab in the left
menu would get this page. The lists are organized in different categories
and we are constantly adding new lists and categories. Let's say we
are interested in gene lists in category ``KEGG'' with keywords
``cell cycle''. Click on the link ``KEGG gene list'' to expand
the search box as shown below. Type cell cycle in the text box and
click submit.
shows the interface to select
a predefined gene list. Clicking on ``Gene List'' tab in the left
menu would get this page. The lists are organized in different categories
and we are constantly adding new lists and categories. Let's say we
are interested in gene lists in category ``KEGG'' with keywords
``cell cycle''. Click on the link ``KEGG gene list'' to expand
the search box as shown below. Type cell cycle in the text box and
click submit.







